Maintaining soil quality is critical to feeding our growing world population. An important and sometimes overlooked aspect of soil is its microbial component. This talk by Dr. Harsh Bais of the University of Delaware highlights the importance of benign soil microbes on plant health.
Author: BUGSS
In Sickness and In Health
Our bodies are home to trillions of microbes and while “germs” are often thought of things to be gotten rid of, most of our microbes don’t make us sick and are even important for our health. In this seminar, Dr. Noel Britton talks about the research into the human microbiome and what we know about the relationship between microbes and human immune system and how this partnership impacts human physiology, from digestion to brain health to drug metabolism. She discussed some of the ways researchers hope to leverage microbes as potential therapeutics for a wide range of health conditions and what you can do to keep your “marriage” with your microbes healthy.
The Story Beyond the Stool Sample
If you’re puzzled by the plethora of information and advertisements about the microbiome, good bacteria, bad bacteria, probiotics and prebiotics, you aren’t alone. This talk answered some common questions about the human microbiome, including: “What even is the human microbiome anyway? Spoiler Alert: it’s more than just bacteria in your poop! How do you measure the microbiome? Are my microbes unique to me? How many microbes are living in and on me and how did they get there? What are these microbes doing for me and why does it matter?
Noel Britton, a postdoctoral researcher at Johns Hopkins University, presented her dissertation work on the human bacterial environment. Throughout the talk, she often referred to the idea of balance. She highlighted that bacteria are not always good or bad and that bacterial species keep one another in check.
The rapid increase in microbiome publications and general research in this field is due to technological advancements. These include DNA sequencing, next-generation sequencing, and pyrosequencing. The avalanche of scientific publications has even made its way into the general person’s daily life through microbiome-focused advertising and products. However, there is still much we have to learn. A microbiome means a collection of microbes and genes that share a common environment. In this presentation, we focused on the human body and its microbiome. The metagenome is the genetic material, while the microbiota refers to the organisms in this environment. Examples of microbiota include bacterial, fungal, and viral species. Microbiota research mainly focuses on bacterial bias rather than fungal or viral organisms.
You may be asking yourself how many microbes are in a person’s microbiome. There are likely more microbes in the human body than there are stars in the Milky Way. The estimated amount of microbes in a single person is one hundred trillion cells and this makes up 57% of the cells in the human body. Various types of microbial quantification methods center around the central dogma. The central dogma is a scientific theory that genetic information flows from DNA to RNA to proteins. As a result, different processes, ranging from microscopy to metabolomics, take advantage of the central dogma to quantify the number of cells in a microbiome. There are different body sample sites used for microbiology sampling like stool, swabs, biopsies, saliva, and urine. For each sample site, there are similar processing steps overall. These steps include DNA extraction, library prep for sequencing, sequencing, and DNA analysis.
In order to extract the DNA from a sample, you must create a solution via vortexing. Next, the solution is centrifuged. This allows for the removal and separation of noncellular debris at the bottom of the centrifuged mix. Researchers continue on working with the supernatant, which is the fluid on top that houses the microbes. The supernatant undergoes high heat exposure and physical beading to break microbe cell walls and extract DNA. Once the DNA is obtained, it is purified and washed for further experimentation.
In preparation for sequencing, adaptation and amplification occur, to ensure there is adequate data for sequencing. During the sequencing process, fragments of the DNA are put together to assemble the entire structure for later data processing. The adapters are removed and the taxonomy of the sample is classified. Similar data are grouped together in an OTU, or operational taxonomic unit. Now that this data is obtained, how can it be analyzed and what questions can it answer? Looking at the data researchers will focus on alpha versus beta diversity. Alpha diversity is the variation within a single sample/group while beta diversity is the dissimilarity between multiple samples/groups. Relative abundance, or the taxonomic composition of each group, is also an important factor for researchers. Researchers also rely on network and correlational analysis to piece together the relationships between the clinical symptoms of their patients along with the samples collected in order to develop a story for their data. In previous research, it has been determined that the phyla makeup mainly remains the same between patients, but the relative abundance and ratios of these groups vary between patients. Even in the case of the McFarlane twins, who are monozygotic twins and probiotic entrepreneurs, they share 100% of their DNA, but only 30% of their gut microbiota. The gut microbiome is a unique identifier between people and there is a possibility of harnessing it as a forensic tool.
Taking a step back, the majority of these studies focus on stool science for gut investigation. Nevertheless, researchers could really sample from anywhere on the human body since microbial communities cover the entire body. Studies depict that there is a similarity between what we commonly think of as good versus bad bacteria. It is truly the amount and ratio of these species and how they interact with one another which leads to beneficial or detrimental effects. Notably, different body sites have their own normal ratios of disease-causing bacteria. For example in the urinary tract, Candida species are normally considered healthy; however, excess amounts can lead to yeast infection. All this information begs the question of how we get our microbes in the first place. They can really come from anywhere. Even a fetus gets microbes from its parent, which go on to influence their immunity early in life. Babies receive additional microbes from the birth canal, breast milk, and their environment in the early stages of life. Microbes are even present on the body after death; though they do decline as the host begins to erode.
Stay tuned for the In Sickness and In Health seminar in which Noel returns to discuss what our microbes do for us and how we can best help them!
Microbes at Work
The last century has witnessed an unprecedented advancement of medical breakthroughs, especially in drug discovery and design. This talk by Dr. Jennifer Kerr of Notre Dame of Maryland University took a historic look at how biopharmaceuticals started, focusing on origin stories of antibiotics and insulin.
Antibiotics were an instrumental discovery in the world of science because they kill bacteria and thus can combat infections. Alexander Fleming is nicknamed the “father of antibiotics”, but there were some key predecessors, like Paul Ehrlich and Gerhard Domagk, who aided in their discovery. Fleming is credited for this discovery due to an accidental finding. One night, he left a petri dish of bacteria open in his lab space. Inadvertently, mold grew and contaminated the sample. Fleming noticed a “zone of inhibition”, or area of no bacterial growth, around where the mold had overtaken. This suggested the mold was creating a substance inhibiting bacterial growth. It did so through a mechanism of action that targets the bacterial cell wall and prevents repair. Fleming ultimately isolated this compound and discovered Penicillin in 1928.
Notably, Penicillin production stalled until the 1940s and there still were hiccups in mass production of the drug. In anticipation of the mass casualties expected in World War II, the UK and USA collaborated on the mass production process and stabilization of Penicillin. Oxford scientists, Dr. Ethel Florey and Margaret Jennings, were instrumental in Penicillin production and purification clinical trials leading up to D-Day. The company Pfizer was involved as well and developed deep citric acid fermentation tanks for large-scale antibiotic production. These initiatives not only aided in the war effort, but also increased antibiotic production and overall yield harvesting substantially. So much so that by 1945 anyone could receive Penicillin, not just the military.
However, antibiotic resistance followed shortly after. Professor Mary Barber was one of the first to detect microbes becoming resistant to Penicillin. Luckily Dr. Selman Waskman and Elizabeth Bugie were hard at work looking for other sources of antibiotics. They systematically screened soil cultures and determined zones of inhibition specifically for pathogenic bacteria and landed upon the antibiotic Streptomycin. Importantly, Streptomycin killed bacteria that were shown to be resistant to Penicillin. The compound was derived from the genus Streptomyces which nowadays supplies half of the global antibiotic supply.
The 1950s became known as the golden age of antibiotic discovery, but unfortunately, scientific advancement has weaned off in recent decades. Antibiotic resistance, on the other hand, has been on the rise, as has antibiotic misuse. In recent years, the WHO has labeled antimicrobial resistance (AMR) an ever-pressing problem for science. Low trial passage, long drug discovery to approval pipeline, and decreased pharmaceutical incentive for profit have hindered development as well. Dr. Kerr also demoed a PEW research model which highlighted the bleak outlook of antibiotic production over the upcoming years.
The seminar switched gears to discuss the role of microbes in another health area. Diabetes occurs when the pancreas cannot create enough insulin to adequately control blood sugar. Sugar in the blood can cause damage across the body in addition to adverse symptoms like frequent urination, excessive thirst, and feeling lethargic. There are variations of the disease. Type 1 diabetics don’t have the necessary cells to produce enough insulin on their own, while Type 2 occurs when the body has developed a resistance to heightened levels of insulin over time.
In the past, Type 1 diabetes was a death sentence early on in life because no one knew the true origin of the disease or how to cure it. Dr. Frederick Banting, John MacLeod, Charles Best, and Gladys Boyd were true pioneers in this regard. In 1921, their research team determined the use of the pancreas in experiments with dogs and began insulin treatment in humans. They found that dogs died shortly after the removal of their pancreas. However, the researchers were still able to keep the animal alive by supplementing it with a pancreatic extract. This pointed them to treatment in human diabetic patients. Nevertheless, insulin harvesting and purification from animal pancreas were extremely inefficient. To get one pound of insulin, the pancreases of 23,500 animals were needed. That totaled 56 million pancreases per year.
It wasn’t until the 1980s that pharmaceuticals searched for a new mechanism of insulin creation using recombinant DNA methods. Recombinant DNA combines DNA between different organisms for expression in a non-originating host species. A modified vector is then put into the host cell. However, the DNA sequence for insulin was not yet discovered, though the 51 amino acid sequence had been determined, courtesy of Dr. Frederick Sanger. Dr. Rosalyn Yalow, Keiichi Itakura, Arthur Riggs, and Herbert Boyer then used immunoassay to detect insulin amounts and reverse engineered the DNA sequence. In addition to DNA expression there was further complex assembly required to fully create active insulin. Finally, Humulin R was developed in 1982. There were further refinements and tweaks to the process which drove down costs.
However, in recent years, different patents and variations of insulin, such as fast/slow acting or different periods of activity, have made insulin prices unattainable for many. This has led to calls for a price cap for insulin and other life-saving drugs. Further discussion occurred at the end of the seminar session regarding what can be done about this. Dr. Kerr suggested getting involved and having general awareness about what’s going on in your community. She also plugged the BUGSS Open Insulin initiative, which aims to create safe insulin as an affordable alternative to what’s on the market. Learn more about Open Insulin at BUGSS here: https://bugssonline.org/
We had a fantastic talk from Dr. Andrea (Andi) Levine, an Assistant Professor of Medicine in the division of Pulmonary & Critical Care Medicine at the University of Maryland School of Medicine. She discussed the current definition of Long-COVID syndrome, what we know about who gets it, why they do, and what we can do to try to both treat and prevent this syndrome.
Long-COVID comes by many names, but refers to symptoms that linger 1-3 months after initial infection, according to the CDC and WHO respectively. We are still very much in this pandemic; Dr. Levine highlighted that at-home testing likely led to the underreporting of overall cases. She highlighted that Long-COVID is its own pandemic and could lead to a mass deterioration event.
Long-COVID symptoms impact almost all organ systems and 80% of patients report at least one symptom that persists long-term. Roughly 50% of patients have ongoing symptoms after 1 month, 5 months, and up to 1 year. The majority of Long-COVID patients were female, obese or with underlying conditions, and around the 50-year age mark. The more symptoms you had earlier on made you more likely to experience Long-COVID symptoms.
The likelihood of a patient acquiring Long-COVID is related to their initial disease severity. A study in The Lancet found that even patients who were less sick initially still reported Long-COVID symptoms. However, that same study alluded that the sicker you were, the more likely you are to experience Long-COVID. The 2-dose vaccination series diminished the likelihood of experiencing persistent illness, but not entirely. Dr. Levine stressed the importance of getting boosters and mask-wearing. Getting vaccinated after COVID-19 infection and long-term symptoms have presented themselves may lead to remission of these symptoms as well. Different COVID variants triggered different persistence in whether patients experienced Long-COVID.
Dr. Levine discussed how the virus can persist in “viral reservoirs” within the body (i.e. the virus can be cleared in a nasal sample, but still present itself in the stool for weeks on end). She stressed again that all organ systems in patients reflect ongoing COVID-19 virus, meaning that no organ tissue is spared. The virus can cross the blood-brain barrier and continue to replicate. In these viral reservoirs (particularly the brain), the virus was seen to mutate from what it was at the time of initial infection. Significant areas of the brain maintained SARS-CoV-2 RNA and structural brain abnormalities could have led to neurological symptoms. COVID-19 infection also caused a prolonged inflammatory state in patients, leading to Long-COVID symptoms. Autoimmunity unmasking by COVID-19 infection is also a probable explanation.
Dr. Levine touched on the mental and social effects of Long-COVID. She discussed how many fear that their experiences are not real and all in their head. Long-COVID correlated with mental health diagnoses and the inability to work. Unfortunately, there are still a lot of unknowns on how inpatient and outpatient therapies impact Long-COVID. She highlighted we, as a society, are at a turning point and must begin to focus on persistent COVID symptoms in addition to the initial infections themselves.
Microbes in Space
The Microbes in Space seminar, co-sponsored by the Maryland Branch of the American Society for Microbiology, highlighted projects conducted in space relating to microbiology. Seminar attendees were joined by Dr. Jennifer Kerr of Notre Dame of Maryland University to present on some key studies happening in space and her own lab’s research.
Kerr first highlighted the many hazards spaceflight has on the human body. These dangers include radiation which damages DNA and lack of gravity which leads to mineral and bone loss. Therefore it is quite astonishing how certain microorganisms can withstand these obstacles, in particular tardigrades. Also known as water bears or moss piglets, tardigrades are tiny and cute invertebrates which prefer to live in water. They are well known for cryptobiosis, Latin for “hidden life”. Cryptobiosis is when there are no signs of metabolic activity, but the organism is still alive. In this state these animals only maintain 0.01% of normal metabolic activity which lets them handle extreme environments. Tardigrades shrivel up and go into what is known as the tun state during this dormancy period. There are also variations of cryptobiosis that tardigrades partake in. For instance, tardigrades in the tun state could survive 125 years without water (anhydrobiosis).
Tardigrade space research began in 2007 with the NASA Foton-M3 Mission studying radiation’s effects on these tiny critters. In a 2019 study, tardigrades in the tun state accidentally crash landed on the moon! Now, is it likely that there is a colony of tardigrades on the moon since there is no water on the moon and those tardigrades arrived in the tun state? Well, a 2021 study investigated tardigrade survival in such high-speed crashes. It was found that moss piglets could survive the impact, but not the shock pressure withstood, so it is unlikely that there is a tardigrade colony on the moon.
Aside from cute water bears, there are also more general microbiome studies happening in space. Specifically, these studies depict how contact surfaces around the International Space Station (ISS) have changed in response to the astronauts who come and go on the Space Station. The microbiomes of crewmembers may influence the microbial composition of ISS habitable surfaces. This is important in managing disease control and preventing contaminants from breaking certain hardware on the ISS. It was found that an astronaut’s microbiome contributes to roughly 55% of the environmental surface microbiome. These findings were not startling, yet it was importantly confirmed that the majority of these were safe and typical bacteria that already exists on the skin. However some were classified as opportunistic pathogens. Opportunistic pathogens have the potential to cause disease, but are unlikely to do so when kept in check by other bacteria or if the person’s immune system is properly functioning. Furthermore, this microbiome snapshot was maintained for a few weeks even after the particular astronaut had left. However this micro-diversity encountered turnover when a new astronaut arrived at the space station and was in constant interplay.
Not only did the microbiomes on ISS surfaces change, even the astronauts who lived there had notable shifts in their own microbial environments. In particular there were 347 bacterial species identified. This varied based on sample sites from the saliva, ears, skin, and nostrils. There were 12 top genera with the highest relative abundance identified across these astronaut samples. Mainly differences were seen in skin samples when astronauts were in flight to and from the Space Station. In the mouth, there were some key, but minimal changes. For instance, saliva had the largest change in composition of bacteria but relative abundance (which is the overall number of bacterial species) stayed the same. One potentially concerning finding with the saliva organisms was that some of them sampled displayed antimicrobial resistant gene markers. The reasoning is unknown, but further research is ongoing.
The NASA Biomedical Engineering for Exploration Space Tech (BEEST) lab is researching health care for exploration. Their goal is to train astronauts in non-invasive treatment of dental cavities. Seminar attendees were also shown a light-hearted video on how astronauts brush their teeth in space. Something as simple as brushing their teeth is even more important in space. There has never been an astronaut who is a dentist, so having preventative care and training is important. Astronauts are even taught techniques up to tooth extraction.
A healthy microbiome is known to be in eubiosis while an unbalanced one is in dysbiosis. For example, the reason that too much sugar leads to tooth decay is because bacteria in the mouth feed on this excess sugar. It causes them to grow more and create more acid. This excess acid leads to a pH shift in which creates a habitable environment for more hardy bacteria to in turn create more acid. This acid also destroys tooth enamel. This leads to cavities and even more body-based diseases. For instance, the dangerous bacteria from the mouth can move through blood to other bodily systems and clog arteries. Maintaining oral hygiene is of utmost priority in space.
This human-microbial research ties back to Dr. Jennifer Kerr’s research at Notre Dame. She is an oral microbiologist and studies teeth in space. Her work centers around Streptococcus mutans. Her lab hopes to help astronauts identify a cavity and use a handheld microwave device to kill the bacteria. Astonighly when this gadget is held to the mouth for only a minute, 99% of the S. mutans are killed. However, in the case of a cavity, demineralization still remains. In order to remineralize it, the astronaut’s body needs to be given the appropriate starting material and the pH has to come back to neutral. This research is still ongoing, but it could have profound impacts, not only on human health in space but even on Earth. According to the Global Burden of Disease Study “oral diseases affect close to 3.5 billion people worldwide” which is why this research and its findings will be so consequential to the world of dentistry and the science and medical communities as a whole.
Co-sponsored by the Maryland Branch of the American Society for Microbiology.

Last fall, with the promise of historic federal and state investments for community recovery, among other important initiatives, Baltimore City announced its Digital Equity Framework – a plan to permanently close its digital divide within the next eight years. Free public Wi-Fi in outdoor community gathering places has been announced as part of the plan, as has connecting our Rec Centers. The grand vision is a municipally owned fiber infrastructure serving all locations in the city. But this ambitious goal won’t be met by technology solutions alone.
Chris Ritzo presented to us about how the city is beginning this work and we discussed the Internet, Wi-Fi, and the power of human networks and community based solutions to combat inequities.
The seminar began with some background information and definitions. The Digital Divide is the gap between those with access to engage online and it disproportionately affects minorities and prevents equal technological access Digital Equity, on the other hand, differs from equality in that it acknowledges systemic barriers in place to hinder others. Lastly Digital Inclusion aims to involve all communities even those most disadvantaged. In order to achieve Digital Inclusion, there are five pillars which must be fulfilled. These include: affordable internet services, access to digital training, quality tech support, access to internet devices, and participation collaboration in the internet sphere. With online school and the overall shift online due to the COVID-19 pandemic, the Divide Divide problem has been further exacerbated and solutions for it become even more pressing.
Throughout the seminar there was active discussion from participants. A question was raised by one attendee on why internet within the City is worse than outside of it, even from the same provider. Another proposed this is because it is harder to lay underground lines in the City. Furthermore, Chris spoke to the disconnect between marketing and engineering sides at companies which also contributes to these problems. Another person highlighted the recent news of a sexual assault that occurred in the Facebook Metaverse. The victim had received responses along the lines of “if you don’t like it then don’t join”. Chris mentioned how this current event ties back to the definition of Digital Inclusion and how moderating community norms is important in addition to creating these novel tools. He also suggested a book, Behind the Screen by Sarah Roberts, identifying the problematic issues of social media. Around the ongoing purchase of Twitter by Elon Musk, Ritzo highlighted Twitter and other social media networks’ claims to support free speech, but how they can never be truly utopian since the judgment lies within the corporation with an end goal of data mining and advertising to its users.
The seminar moved on to discuss technical aspects of getting access to the Internet. For instance, a router helps us connect wired/wirelessly to a laptop onto the Internet. This router also protects us and our data to some extent via firewall. First Mile is the idea of democratizing internet setup. In this concept, individuals can also set up Wi-Fi services, not just big corporations. The seminar also touched upon running speed tests. Chris said speed is important, but not the only thing that matters. It was recommended to survey nearby Wi-Fi channels, buy an extender, and understand the channel overlaps of your current network and where it should be. There were also online sites provided to see how crowded the channels are in your neighborhood. In the same vein, conduct basic latency tests, in particular latency under load also known as bufferbloat, to determine where your internet stands as it buffers in the case there is too much data.
Overall, data mechanics surrounding internet accessibility maintains a key driver in creating a community based solution to this problem. Science and technology will benefit greatly when there is contribution and inclusion in which there is equitable and diverse representation across the Internet.
Go with Your Guts!
Go with your guts, and the billions of bacteria that are in them!
Test your own personal microbiome (or your pet’s) –
In this class you will be able to investigate your own gut microbiome, or the microbes of your pets, roommates, or family members. Our microbiome (the microbes living in and on our bodies) are believed to have profound effects on our immune systems, health, and susceptibility to disease. Find out what you can learn about your own health and wellness. In this class you’ll perform lab work to isolate the microbes present, sequence and identify those microbes, and then learn what the results mean for YOU personally.
In this 3 Saturday class you’ll be able to design your own experiment to compare any two samples! What samples do you want to compare? How many gut microbes you share with your dog? What about with another human? Does your gut microbiome change with your diet? What if you ate pizza for two weeks straight? (We do not endorse pizza as a sole source of nutrition!)
Isolate and process DNA from your samples. Use the polymerase chain reaction (PCR) to amplify the DNA of the microbes. Determine the different species of in your two samples. For those interested in programming and computational biology, the entire class will learn how to analyze the sequencing data and perform comparative analysis to uncover further information (don’t worry, this class is appropriate for beginners!).
Everyone gets two biological samples to test, here are some experimental suggestions. If none of these interest you, just ask us if you have another idea.
Some experimental suggestions:
1) Test your microbiome vs. your roommate (or friend, or family member)’s microbiome *
2) Test your microbiome vs. your pet **
3) Test your microbiome before and after a lifestyle change such as a change in diet, exercise or sleep habits
4) Test your pet microbiome before and after a change in your pet’s lifestyle
* Mandatory: you must get their consent
**Well, at least ask?
Computational Modeling with R
Week 1: Deterministic Modeling
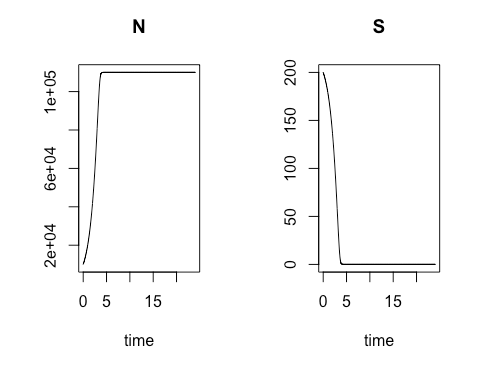
In the opening session March 2, after guiding class members through the installation of R and the RStudio integrated development environment on their laptops, Ms. Mbuguiro presented an introduction to deterministic modeling. Each model considered was a mathematical explanation of a biological process of interest. “Deterministic” means that the output of the model depends solely on the precise values and conditions used as input for the model, and not on any variables that may have a random or other probabilistic distribution. The class recreated deterministic models – expressed in R – for drug delivery via nanoparticles and for bacteria grown in culture. We also explored fitting functions to our data – that is, optimizing a model to best account for our data – using the least squares method. Rather than writing code from scratch, we began by modifying short segments of existing code provided in the development environment. This enabled us to avoid trivial mistakes and permitted the focus to remain on gaining experience using mathematical concepts expressed in R to study a biological system.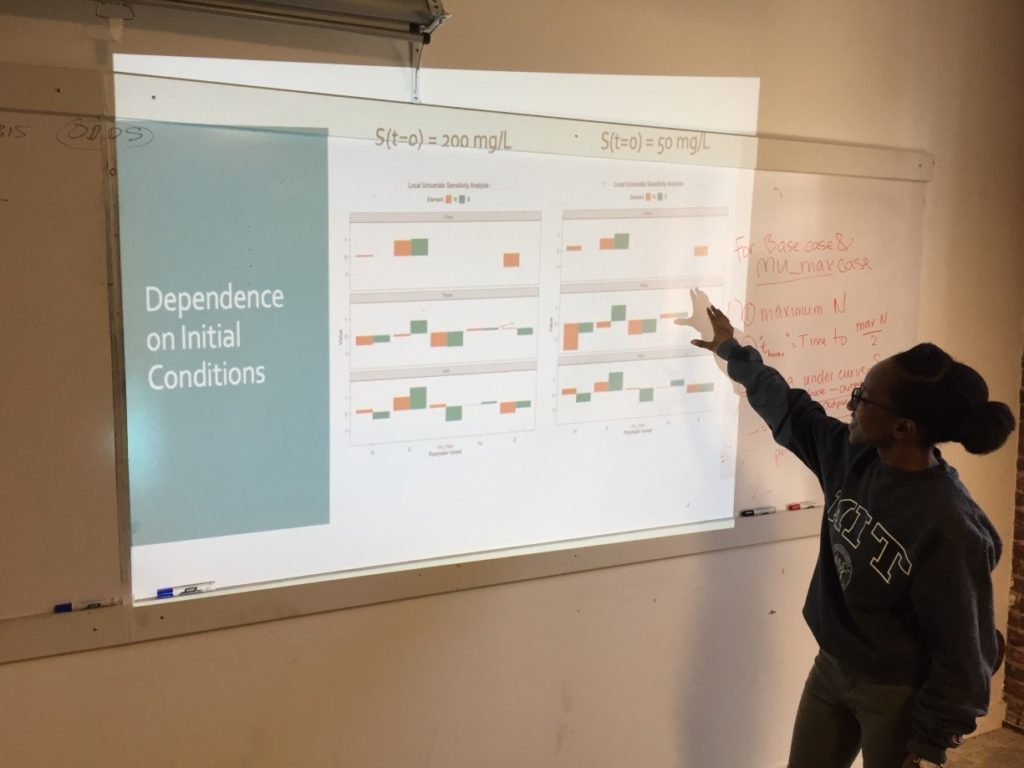
Week 2: Modeling growth rates
On the second Saturday, March 9, we explored using R for a common laboratory task: calculating the varying rate of change over time for a biological process involving a material of interest, and using the results to obtain a close estimate of the quantity of that material which is present at various time intervals. The varying rates of change are values of an ordinary differential equation. Using these values to obtain numerical approximations of the concentration of the material of interest at different points in time can be especially useful when direct analytic evaluation is difficult. In particular, the class focused on modeling the growth rate of bacterial colonies. We started with the essential relationship between the concentration of bacterial cells present and the growth rate of that cell concentration when there are no external constraints, such as limits on food supply. By calculating the rate of change in cell growth over very small changes in time using a species-specific growth rate constant and the concentration at the start of the time period, we were able to estimate the cell concentration at each incremental time point. When plotted, these cell concentration calculations formed a smooth curve that revealed exponential growth over the time sequence.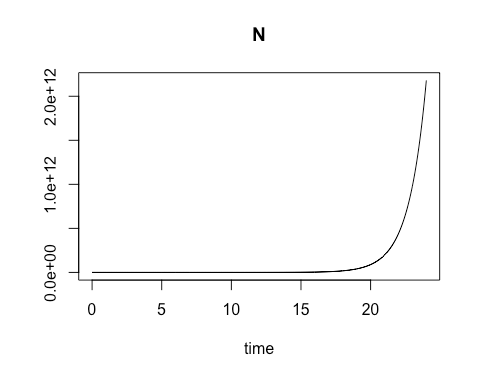
Week 3: Sensitivity Analysis
On the third Saturday, March 16, we conducted a sensitivity analysis on the Monod model of cell concentration increase and substrate depletion. Creating code in R for changing – one at a time –each relevant initial condition of the system and each rate-governing parameter, we explored the effect on the final outputs – cell concentration and substrate concentration – of a 10% change in each of the initial conditions and parameters. Looking at the effect of a 10% increase in the growth constant, we learned how to get a close estimate of the time required to reach half of the maximum cell concentration. As an illustration of how easily R can accommodate new functions to meet special needs, Ms. Mbuguiro wrote a “helper function” to find the position number within a sequence of time values of the particular value associated with a cell concentration that had reached 50% of the maximum. The calculation of all output changes driven by an incremental change of one input parameter is called a univariate sensitivity analysis. Extending our exploration of R for standard statistical manipulations, we normalized the outputs of the entire sensitivity analysis – that is, we converted the change in each output from an absolute measure into a measure that is relative to the 10% change of the growth constant. As a last step, the class wrote the R code to create a graphical representation of the normalized output for multiple univariate sensitivity analyses, showing the effect of 10% changes in various parameters, considered one at a time, upon properties associated with cell concentration (N) and with substrate concentration (S). These properties include: Cmax, the maximum cell or substrate concentration; Thmax, the time required to arrive at one half of Cmax; and the Area under Curve (AUC), a measure of concentration over time that can be used to calculate average concentration during the time period. The area under the curve for the substrate concentration is often used in drug development research as a measure of “exposure” to the substrate. As an aid to visualization, the class made use of another R “helper function” contributed by Ms. Mbuguiro called output_calculator2, which works in concert with other R functions to produce the final output.
About the Instructor
Wangui Mbuguiro is a Ph.D. candidate in the Biomedical Engineering Program at Johns Hopkins. Her research and passions center on engineering tools to better understand and treat menstrual disorders as part of the Computational Design of Therapeutics Lab. Outside of lab, Wangui enjoys encouraging scientific inquisition as an instructor and mentor at the Baltimore Underground Science Space, as well as building opportunities and community for underrepresented students in STEM at Johns Hopkins. Lastly, Wangui is a MIT alumna (B.S., Bioengineering, 2017), National Science Foundation Fellow, and friendly neighborhood scientist. You can connect with her on twitter (@WanguiMbuguiro) or LinkedIn.[1] Schoeberl, B. et al., Systems biology driving drug development: from design to the clinical testing of the anti-ErbB3 antibody seribantumab (MM-121). npj Systems Biology and Applications (2017) 3, 16034; doi:10.1038/npjsba.2016.34; published online 5 January 2017. [2] Hosseini, I. and Mac Gabhann, F., Mechanistic Models Predict Efficacy of CCR5-Deficient Stem Cell Transplants in HIV Patient Populations. CPT Pharmacometrics Syst. Pharmacol. (2016) 5, 82–90; doi:10.1002/psp4.12059; published online 16 February 2016. [3] Maier, R.M., Bacterial Growth. In Environmental Microbiology (Maier, R.M., Pepper, I.L., and Gerba, C.P., eds., 2nd ed., Academic Press, 2009), Ch. 3, p. 37-54. https://doi.org/10.1016/B978-0-12-370519-8.00003-1. (http://www.sciencedirect.com/science/article/pii/B9780123705198000031)
City Nature Challenge


CNC is a platform that brings together nature enthusiasts from around the globe.
Have you ever wondered-
What is that peculiar bug chilling out on my plant called? I planted tomatoes, but not this other plant. What is that? A colorful bird loves to stop by my bird feeder. I wonder what my new friend’s name is? If you answered yes to any (or all) of these questions, CNC is your chance to find some answers and to contribute to science while doing that.
Want more information?
Visit the City Nature Challenge site